Check out the latest publication from the Mehl/Cooley Lab!
Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity
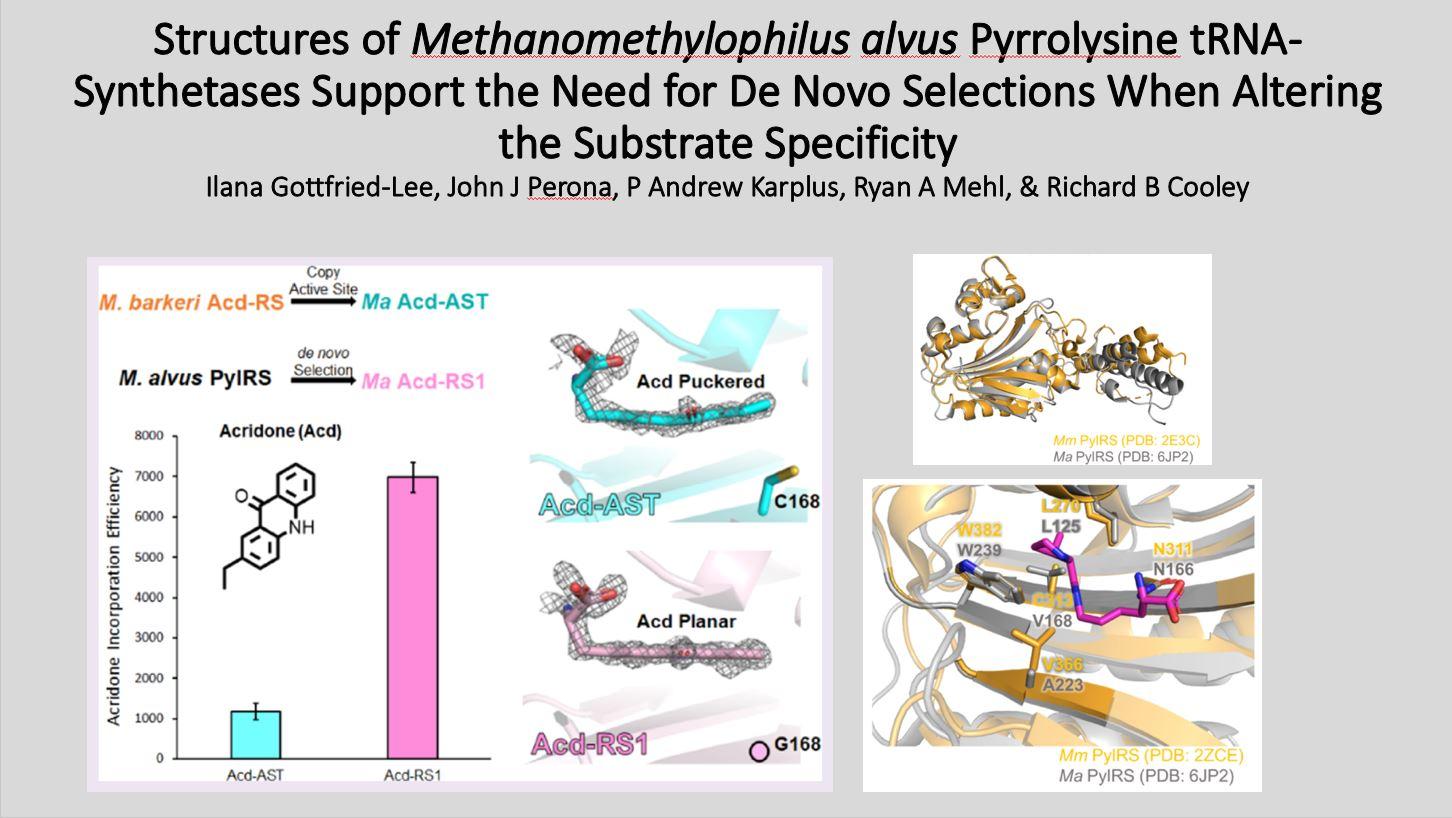
We recently created two new Ma PylRS variants for the incorporation of the fluorescent amino acid, acridonyl-alanine (Acd), into proteins at amber codons: one based on “transplanting” active site mutations from an established highefficiency Mb PylRS and one that was de novo selected from a library of mutants. Here, we present the crystal structures of these two Ma PylRS variants with Acd/ATP bound to understand why the “active site transplant” variant (Acd-AST) displayed 6-fold worse Acd incorporation efficiency than the de novo selected PylRS (called Acd-RS1).